Project One
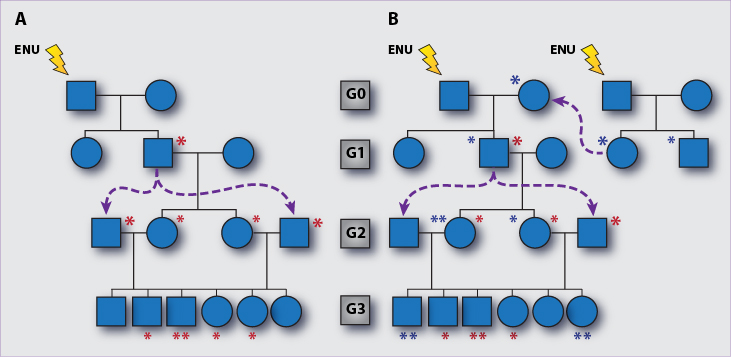
Forward genetic analysis begins with mutagenesis using the alkylating agent N-ethyl-N-nitrosourea (ENU), which is administered to male C57BL/6J mice to create germline point mutations. ENU eliminates most spermatogonia in these Generation 0 (G0) animals, causing transient sterility. Between 10 and 100 precursors, each harboring approximately 12,000 point mutations, repopulate the testis over a period of 12 weeks following ENU administration. About 6,000 mutations are incorporated into each gamete. These mutations are transmitted to G1 offspring in a heterozygous state, and brought to homozygosity in G3 mice using one of two different inbreeding strategies (Figure 1). It is estimated that about 11 coding changes are brought to homozygosity in each G3 mouse. Currently we are generating 10 G2 females to back-cross with the G1 male to generate 50 G3 mice per pedigree, covering all mutations in the homozygous state if they are viable as such.
DNA is obtained from each G1 male, extracted, fragmented and exome enriched for whole exome sequencing. We sequence 80 G1 exomes in parallel every 12 days. To identify ENU-induced mutations, the reads are mapped to the mm10/GrCM38 genome using BWA and variants are called using SAMtools. These variants are then annotated and filtered to exclude non-coding and non-splicing associated variations, as well as variations appearing in multiple unrelated G1 samples. Finally the remaining variants are filtered for quality and uploaded into Mutagenetix. Variants are annotated according to their inferred impact on the protein, assessed using the program Polyphen-2. From these final remaining variants a run file is created for Ampliseq panels, which are used to amplify every mutation site in every mouse in the pedigree (i.e., the G1 and all its descendants). Genotyping is accomplished by Ion Torrent sequencing of bar-coded DNA libraries. The genotyping data are stored on Mutagenetix until such time as phenotypic screening is complete.
All G3 mice are screened for phenovariance using more than 40 distinct assays, and laboratory personnel are alert to the discovery of visible or behavioral phenotypes as well. Phenotypes of interest include abnormal responses to sub-lethal inocula of common pathogens, decreased or elevated levels of immune cells, abnormal GI homeostasis when challenged with DSS, and abnormalities of the innate or adaptive immune response. Declaration of a significant phenotype was previously dependent on human interpretation of screening data, but we are now moving toward a statistically-based analysis of phenotypic scores to isolate significant variations and the detection of linkage. Once phenotypic data are available, the statistical algorithm correlates the phenotypic scores and genotypes of the G2 and G3 mice in order to determine the causative mutation (or mutations) contributing to the observed phenotype.
After specific genes are implicated by this instantaneous form of linkage mapping, they are further validated by the creation of knock-in or knockout mice via gene targeting. Recently several genome-editing technologies, such as Zinc-Finger Nuclease (ZFN), Transcription Activator-Like Effector Nucleases (TALENs) and the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated 9 (Cas9) system, have been successfully used to produce genetically modified mice. Among these, CRISPR/Cas9 system is the most popular genome-editing technology due to its ease of manipulation and high efficiency.