Project Three
Systems Biology Analysis of Innate Immune Responses
The innate immune system is critical for host defense but when unchecked or dysregulated, can cause severe inflammatory disease. The innate immune response is therefore regulated at multiple levels. Signaling networks initiated by the engagement of a variety of pattern recognition receptors (e.g. Toll-like receptors, Nod-like receptors) can be modulated by mechanisms such as feedback and crosstalk among the signaling pathways.
Currently, many therapeutics that modulate innate immunity act broadly to enhance or repress responses. For instance, many vaccine adjuvants under development are TLR agonists and therefore activate the entire signaling pathway. Conversely, many anti-inflammatory treatments broadly suppress immune responses, rendering the patient vulnerable to infection. We employ systems biology techniques to generate and analyze unbiased global datasets to reveal previously unknown regulators of the innate immune response to guide the development of more focused and precise therapeutic interventions.
The goal of this project is to characterize the role that genes and proteins identified through network analysis play in the innate immune response in macrophages and dendritic cells. We will pursue genes that have an ENU-induced protein-coding mutation where this mutation causes a transcriptional phenotype. We are focusing on those genes for which there is little prior knowledge of their function in innate immunity.
Aim 1. Determine the mechanism by which the gene of interest modulates, directly or indirectly, transcriptional responses in innate immune cells.
This project will use high throughput global measurements to evaluate the precise effect of mutations in our genes of interest on transcriptional networks in vitro. Transcriptomic analysis will rapidly contextualize genes of interest in known innate immune pathways and allow us to refine these pathways. Genome-wide location analysis will be used to determine the mechanism by which genes regulate transcription. Lastly, mass-spectrometry approaches will be used to determine how genes of interest control enhanceosome formation at the promoters of innate immune response genes.
Aim 2. Delineate the role of the gene of interest macrophage and dendritic cell signaling networks.
Proteins function in networks that include numerous protein-protein interactions and often multi-protein complexes. We will use mass spectrometry based proteomic techniques to determine the interactomes of selected genes with ENU mutations and the roles of genes of interest on signaling pathways and on the production of lipid immune mediators. We will also employ mass cytometry (CyTOF) to determine the effect of genes of interest on signaling pathways.
Aim 3. Determine the role of selected genes in controlling cellular and in vivo immune responses to relevant NIAID Category A-C Priority Pathogens.
We will determine the role of selected genes in macrophage cellular and in vivo responses to L. monocytogenes and S. typhimurium as well as the roles of genes of interest in dendritic cell responses to influenza will be determined in vitro and in vivo.
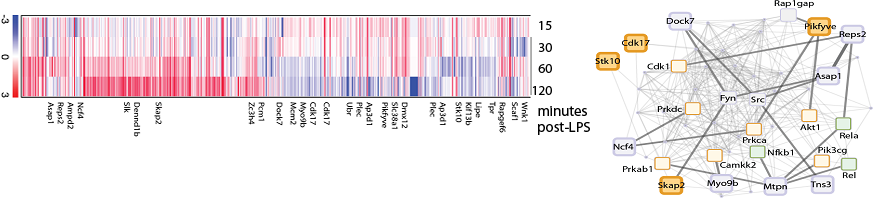
Left: Time-course expression of phosphorylated proteins in macrophages following LPS stimulation. Red (blue) indicates increased (decreased) abundance of the phosphorylated species at the indicated time point. Data is presented as log2 ratios relative to T=0. A subset of ~6000 phospho peptides covering ~2200 proteins is shown.
Right: Cytoscape visualization of a network constructed from nearest-neighbor interactions of all phospho-proteins identified that have incidental ENU-induced mutations in the Mutagenetix database (large squares). Selected interacting proteins with known functions in macrophages are labeled (small squares). Kinases are labeled orange and transcription factors are labeled green.