The Beutler and Goodnow team will produce an unparalleled resource of mouse mutations for experimental and systems analysis of innate and adaptive immunity.
Links
ENU-induced Single Nucleotide Variants (SNV)
The Genetics Core produces a diverse experimental mouse resource of missense gene variants, where that genetic variation is expressed on an otherwise inbred strain background and where the tools of experimental biology can be applied to full effect. Many missense mutations produced by ENU yield unique counterparts of human variation, illustrated for example by analysis of the effects of quantitative variants in Zap-70, Ligase 4 and Ikappa kinase gamma.
Single Nucleotide Variants (SNVs) are introduced into C57BL/6 mouse spermatagonial stem cells with the chemical mutagen N-ethyl-N-nitrosourea (ENU). The chemical is an alkylating agent and has preference for A/T to T/A base transversions and A/T to G/C base transitions. ENU is administered to male mice in three weekly doses of 100mg/kg body weight. After a period of transient sterility, the mice regain fertility and transmit mutations to their offspring at a rate of about 1 bp change per million bp genomic DNA; hence each G1 mouse is heterozygous for approximately 3,000 point mutations with around 35-75 coding/splice mutations per G1 mouse. Genomic DNA from each G1 mouse is extracted from blood, sheared, and exonic DNA fragments captured with Agilent mouse exome capture kits for whole exome sequencing. Exome capture followed by sequencing reliably identifies the rare, ENU-induced, de novo mutations in C57BL/6 mice (>98% validate). In addition to generating null mutations, the ENU approach allows the analysis of more subtle variations such as hypomorphic, dominant-negative, or gain of function mutations that provide mechanistic information relevant to understanding human disease SNVs.

Exome sequencing pipeline to identify SNVs
The bioinformatics workflow holds together a number of open-source analysis tools and employs a Perl code-base to perform custom filtering, reporting, and job process control. The bioinformatics pipeline for SNV and indel identification uses the ENSEMBL gene sequence for humans and mice to map the data.
Access to the Missense Mutation Library via Website
The annotated SNVs are listed on both UTSW and APF/ANU databases- see links above. Researchers can register to obtain notification of mutations in genes of interest using the Genewatch list on APF database.
Annotation of SNVs for informative mouse model selection
As G1 mice are subjected to whole-exome sequencing, large numbers of ENU-induced mutations are detected within and near coding exons. However, as >98% accuracy has been achieved validation is only performed when the SNV is requested for propagation. Validated or not, mutations are computationally analyzed and those that cause coding change or occupy splice junctions (defined as nucleotides within 10 residues of the exon boundary) are flagged for export to the Missense Mutation Library. The quality of the SNV call can be determined by read depth and allele frequency information.
Each SNV is given a unique identification number. The gene, chromosome and exact location (Ensembl coordinates), amino acid change or distance from the splice junction, Polyphen-2 score, SIFT analysis are listed in sortable columns. All missense mutations are analyzed using Polyphen-2 (altered to use mouse sequence databases) to predict the effect of the variation on the protein. Polyphen assigns a high score (greater than 0.95) of probably damaging and an intermediate to high (0.44-0.95) of possibly damaging. In a retrospective analysis of causal SNVs, 75% of SNVs with a phenotype have been scored by polyphen2 as probably damaging.
A computational pipeline adds further annotation, including links to Mouse Genome Informatics (MGI), Human Rare diseases (Orphanet), and gene expression links to Immgen. SNV availability indicates whether the SNV is in a mouse that is cryopreserved or still breeding in the facility.
ENU Mouse Pedigrees for Gene Discovery
Male C57BL/6 mice are treated with three doses of ENU 100mg/Kg over 3 weeks. Following resting for 8 weeks these male mice, G0, are set up with female mice that carry mutations derived from unrelated ENU mutagenized males (G0′) or C57BL/6 females. Unrelated G1 mice are paired to start a pedigree and produce G2 male and female mice G2 x G2 brother-sister mating are set up to produce G3 mice or G2 female daughters are cycled past their G1 father to produce G3 mice for phenotypic screening.
For each pedigree this includes cycling three G2 daughters past their G1 father or setting up three G2 x G2 brother-sister matings per pedigree. If a phenotype of interest is found within the G3 pedigree, the pedigree will be expanded for progeny testing to confirm heritability and confirmation of phenotype.
At this stage, the pedigree will be referenced back to the baseline G1 sequence to look at the SNVs to genotype for the causal SNV in affected mice. Sperm and DNA from mice from each pedigree are cryopreserved.
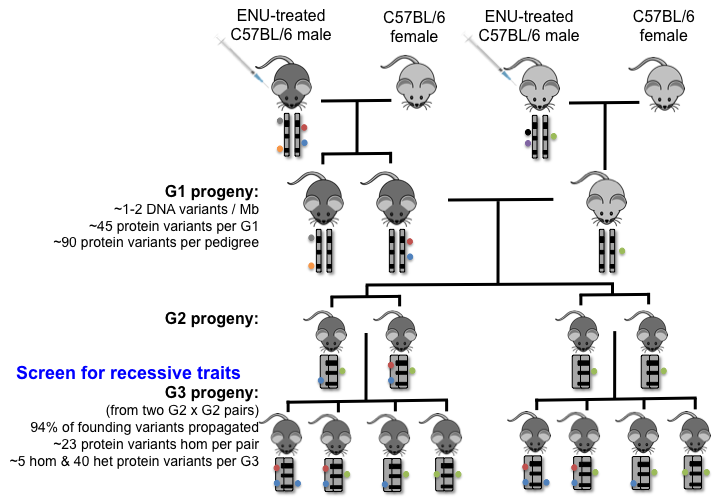
Representative ENU G2 intercross pedigree founded by two unrelated G1 mice.