The Annotation Core is responsible for mutation annotation and archiving. It will also continue to develop software to inform all users and the scientific community of:
- mutant mice that are available (including those known to have a phenotype and those that have “incidental” mutations)
- the documented or predicted effects of these mutations on the mice
- the procedure by which these mice can be obtained.
All data developed by Annotation Core will be written to the Mutagenetix relational database, an integrated, searchable resource for ENU-induced mouse phenotypes and mutations.
Automated annotation of phenotypic mutations includes an appraisal of the likelihood that each mutation will cause damage to the affected protein, performed by local implementation of Polyphen-2, a program that relies on multiple indices including high-quality multiple sequence alignment, probabilistic calculation based on a machine-learning method, and domain structure to predict whether variants from the WT sequence are damaging
All missense and putative splicing mutations are examined by SMART (Simple Modular Architecture Research Tool), run as a remote batch program. The domain structure is drawn automatically, so that a “mouse-over” illustration of each protein is rendered. The location of the mutation within the protein sequence is indicated by a vertical red line. Links to domain definitions can be accessed by clicking on particular domains.
Each page has an annotated review of the mutation and gene in question, illustrated in detail, and often containing many external links to relevant databases detailing human diseases, other mouse alleles, and structural data.
Added informatics capabilities, to be written in the course of the funding period, will include assessments of where individual genes are expressed, the effects of immune activation on expression of individual genes, and the transcriptional regulatory influences to which individual genes are subject.
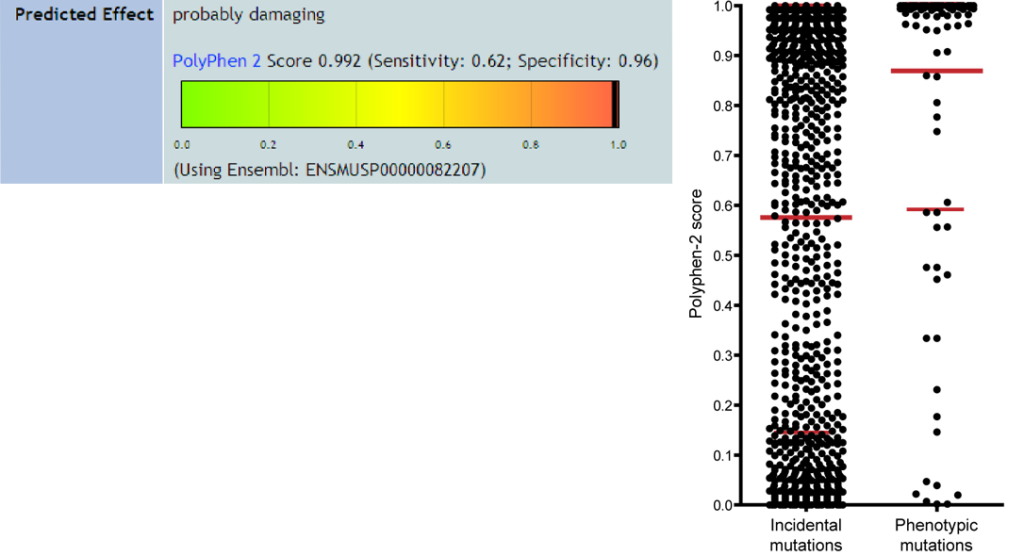
Left: Prediction of the effect of the L499P mutation in Tlr9 from the record for CpG1.
Right: Polyphen-2 scores for phenotypic mutations are on average higher than those of incidental mutations, indicative of more damaging effects (P<0.0001, Mann-Whitney test). Each point represents and individual mutation. N=1548 for incidental mutations; N=127 for phenotypic mutations. Error bars represent one standard deviation.